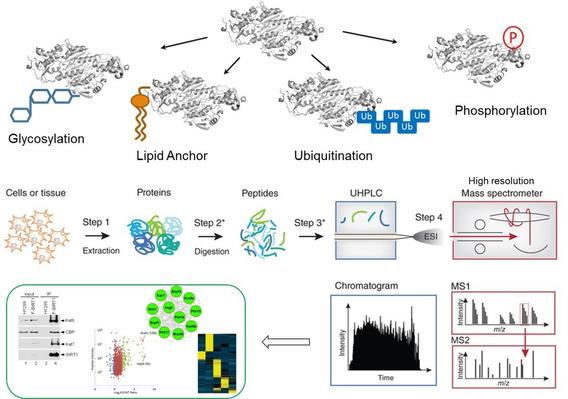
Post-translational modifications of proteins (PTMs) profoundly impact the substrates’ structure, activity and protein-protein interactions. Dysregulation of the PTM pathways has been widely implicated in a variety of cellular processes and diseases including cancer, aging and metabolic diseases. Over 200 different types of PTMs have been identified and yet, functional significance of these PTM pathways in cells and disease progression remain largely unexplored. A key challenge is the lack of effective technologies to comprehensively characterize the PTM proteome and accurately quantify the site-specific dynamics of PTM targets in cellular processes. The Chen lab develops functional proteomics technologies to systematically identify novel PTM regulatory targets and discover aberrantly regulated PTM pathways in different disease states.
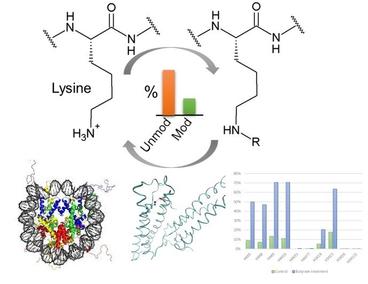
1. Quantitative chemical proteomics technology to study lysine modification pathways.
Lysine is an important target of protein modifications playing key roles in the epigenetic regulation, cell signaling and protein degradation. Lysine acetylation is an ancient protein modification first discovered in 1963. Recently, the MS-based global analysis of lysine acetylation proteome revealed multi-faceted roles of lysine acetylation in regulating gene transcription, enzyme activity and cellular metabolism. Based on the success of this technology platform, we are combining quantitative proteomics technology to identify lysine acetylation targets of the regulatory enzymes or the dynamics of lysine acetylation proteome in different disease states. We are also developing new chemical proteomics strategies to determine absolute site-specific lysine acetylation stoichiometry in cells and tissues and investigate the roles of lysine acetylation in protein homeostasis.
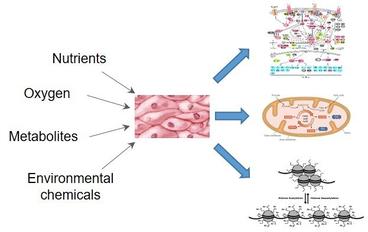
2. Environmental impacts on cell signaling and epigenetics.
Changes in the cell microenvironment including oxygen availability, nutrient concentrations and the existence of the chemically active compounds may dramatically alter the cellular metabolism, epigenetic regulation and signaling pathways. Such processes have been widely implicated in cancer initiation and proliferation. During these processes, cells activate specific signaling cascades through posttranslational modifications which eventually lead to the epigenetic reprogramming of the gene transcriptions. Understanding the dynamics of these signaling events is critical to discover novel regulatory mechanisms and pathways that are functionally essential in these processes. We are applying the MS-based quantitative proteomics technology to systematically identify the temporal and spatial dynamics of the signaling networks in cancer cells in response to the environmental perturbations with an ultimate goal to reveal novel targets for therapeutic intervention of tumorigenesis and tumor progression.